| 40 |
DeviceEditor format (.xml) design files |
| DeviceEditor design files are based upon XML, and contain all of the information required to fully recapitulate designs on the DeviceEditor canvas. Third-party bioCAD tools can output DeviceEditor design files for integration with DeviceEditor and thereby j5 DNA assembly design automation. Here is the XML schema for DeviceEditor format design files: (design.xsd) Here is an example DeviceEditor design: (DeviceEditor_example.xml) 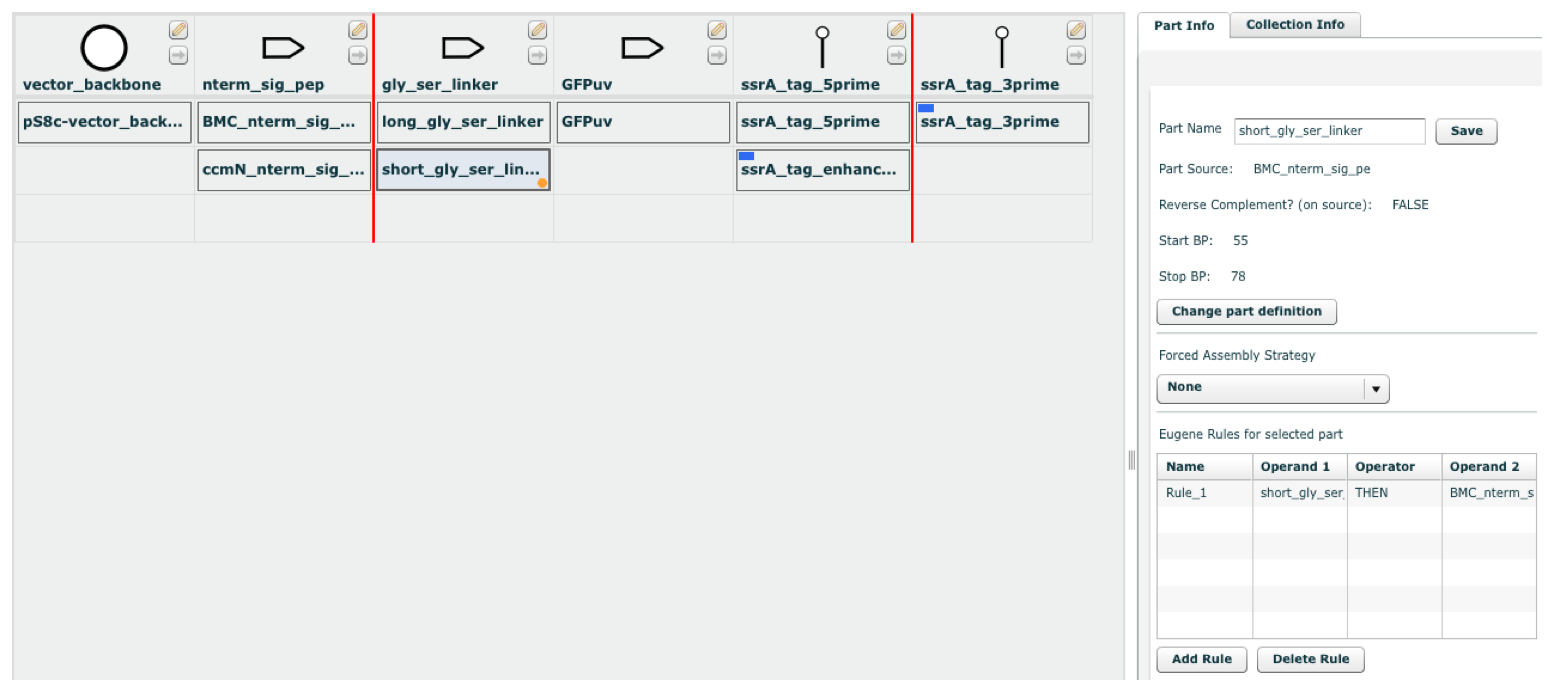 Several key aspects of this example design to highlight are: 1) This is a combinatorial design, so there are multiple parts (e.g. "BMC_nterm_sig_pep" and "ccmN_nterm_sig_pep") in the same collection column (e.g. "nterm_sig_pep"). 2) A part has a Eugene design specification rule associated with it ("short_gly_ser_linker"). 3) Forced assembly strategies have been set for several parts (e.g. "ssrA_tag_3prime"). 4) Direct synthesis firewalls and forced relative overhang directives have been set for several collection columns (e.g. a direct synthesis firewall for column "nterm_sig_pep", and a +2 forced relative overhang for column "vector_backbone"). Here is the stylized DeviceEditor design file for the above example: <?xml version="1.0" encoding="UTF-8"?> <de:design xsi:schemaLocation="http://jbei.org/device_editor design.xsd" xmlns:de="http://jbei.org/device_editor" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"> <de:version>4.4</de:version> <de:sequenceFiles> <de:sequenceFile id="1383621055622750"> <de:format>Genbank</de:format> <de:content><![CDATA[LOCUS pj5_00001 5299 bp dna circular UNK 26-OCT-2009 DEFINITION promoter seq from pBAD33. ACCESSION unknown KEYWORDS ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; . FEATURES Location/Qualifiers CDS complement(7..885) /label=araC protein_bind 914..931 /label=operator\O2 promoter complement(1036..1064) /label=araC\promoter protein_bind 1072..1093 /label=operator\O1 misc_binding 1115..1128 /label=CAP\site protein_bind 1124..1162 /label=Operator\I2\and\I1 promoter 1161..1188 /label=pBAD\promoter RBS 1216..1235 /label=RBS CDS 1236..2018 /vntifkey="4" /label=GFPuv misc_feature 1661..1661 /label=XhoI_silent_mutation misc_feature 1760..1760 /label=BamHI_silent_mutation CDS 1953..2015 /vntifkey="4" /label=signal_peptide terminator 2034..2162 /label=dbl\term rep_origin complement(2164..4392) /label=pSC101** terminator 4393..4498 /label=T0 misc_marker complement(4514..5173) /label=CmR ORIGIN 1 gacgtcttat gacaacttga cggctacatc attcactttt tcttcacaac cggcacggaa 61 ctcgctcggg ctggccccgg tgcatttttt aaatacccgc gagaaataga gttgatcgtc 121 aaaaccaaca ttgcgaccga cggtggcgat aggcatccgg gtggtgctca aaagcagctt 181 cgcctggctg atacgttggt cctcgcgcca gcttaagacg ctaatcccta actgctggcg 241 gaaaagatgt gacagacgcg acggcgacaa gcaaacatgc tgtgcgacgc tggcgatatc 301 aaaattgctg tctgccaggt gatcgctgat gtactgacaa gcctcgcgta cccgattatc 361 catcggtgga tggagcgact cgttaatcgc ttccatgcgc cgcagtaaca attgctcaag 421 cagatttatc gccagcagct ccgaatagcg cccttcccct tgcccggcgt taatgatttg 481 cccaaacagg tcgctgaaat gcggctggtg cgcttcatcc gggcgaaaga accccgtatt 541 ggcaaatatt gacggccagt taagccattc atgccagtag gcgcgcggac gaaagtaaac 601 ccactggtga taccattcgc gagcctccgg atgacgaccg tagtgatgaa tctctcctgg 661 cgggaacagc aaaatatcac ccggtcggca aacaaattct cgtccctgat ttttcaccac 721 cccctgaccg cgaatggtga gattgagaat ataacctttc attcccagcg gtcggtcgat 781 aaaaaaatcg agataaccgt tggcctcaat cggcgttaaa cccgccacca gatgggcatt 841 aaacgagtat cccggcagca ggggatcatt ttgcgcttca gccatacttt tcatactccc 901 gccattcaga gaagaaacca attgtccata ttgcatcaga cattgccgtc actgcgtctt 961 ttactggctc ttctcgctaa ccaaaccggt aaccccgctt attaaaagca ttctgtaaca 1021 aagcgggacc aaagccatga caaaaacgcg taacaaaagt gtctataatc acggcagaaa 1081 agtccacatt gattatttgc acggcgtcac actttgctat gccatagcat ttttatccat 1141 aagattagcg gattctacct gacgcttttt atcgcaactc tctactgttt ctccataccc 1201 gtttttttgg gaatttttaa gaaggagata tacatatgag taaaggagaa gaacttttca 1261 ctggagttgt cccaattctt gttgaattag atggtgatgt taatgggcac aaattttctg 1321 tcagtggaga gggtgaaggt gatgcaacat acggaaaact tacccttaaa tttatttgca 1381 ctactggaaa actacctgtt ccatggccaa cacttgtcac tactttctct tatggtgttc 1441 aatgcttttc ccgttatccg gatcatatga aacggcatga ctttttcaag agtgccatgc 1501 ccgaaggtta tgtacaggaa cgcactatat ctttcaaaga tgacgggaac tacaagacgc 1561 gtgctgaagt caagtttgaa ggtgataccc ttgttaatcg tatcgagtta aaaggtattg 1621 attttaaaga agatggaaac attctcggac acaaactcga atacaactat aactcacaca 1681 atgtatacat cacggcagac aaacaaaaga atggaatcaa agctaacttc aaaattcgcc 1741 acaacattga agatggatct gttcaactag cagaccatta tcaacaaaat actccaattg 1801 gcgatggccc tgtcctttta ccagacaacc attacctgtc gacacaatct gccctttcga 1861 aagatcccaa cgaaaagcgt gaccacatgg tccttcttga gtttgtaact gctgctggga 1921 ttacacatgg catggatgag ctcggcggcg gcggcagcaa ggtctacggc aaggaacagt 1981 ttttgcggat gcgccagagc atgttccccg atcgctaaat cgagtaagga tctccaggca 2041 tcaaataaaa cgaaaggctc agtcgaaaga ctgggccttt cgttttatct gttgtttgtc 2101 ggtgaacgct ctctactaga gtcacactgg ctcaccttcg ggtgggcctt tctgcgttta 2161 tacctagggt acgggttttg ctgcccgcaa acgggctgtt ctggtgttgc tagtttgtta 2221 tcagaatcgc agatccggct tcagccggtt tgccggctga aagcgctatt tcttccagaa 2281 ttgccatgat tttttcccca cgggaggcgt cactggctcc cgtgttgtcg gcagctttga 2341 ttcgataagc agcatcgcct gtttcaggct gtctatgtgt gactgttgag ctgtaacaag 2401 ttgtctcagg tgttcaattt catgttctag ttgctttgtt ttactggttt cacctgttct 2461 attaggtgtt acatgctgtt catctgttac attgtcgatc tgttcatggt gaacagcttt 2521 gaatgcacca aaaactcgta aaagctctga tgtatctatc ttttttacac cgttttcatc 2581 tgtgcatatg gacagttttc cctttgatat gtaacggtga acagttgttc tacttttgtt 2641 tgttagtctt gatgcttcac tgatagatac aagagccata agaacctcag atccttccgt 2701 atttagccag tatgttctct agtgtggttc gttgtttttg cgtgagccat gagaacgaac 2761 cattgagatc atacttactt tgcatgtcac tcaaaaattt tgcctcaaaa ctggtgagct 2821 gaatttttgc agttaaagca tcgtgtagtg tttttcttag tccgttatgt aggtaggaat 2881 ctgatgtaat ggttgttggt attttgtcac cattcatttt tatctggttg ttctcaagtt 2941 cggttacgag atccatttgt ctatctagtt caacttggaa aatcaacgta tcagtcgggc 3001 ggcctcgctt atcaaccacc aatttcatat tgctgtaagt gtttaaatct ttacttattg 3061 gtttcaaaac ccattggtta agccttttaa actcatggta gttattttca agcattaaca 3121 tgaacttaaa ttcatcaagg ctaatctcta tatttgcctt gtgagttttc ttttgtgtta 3181 gttcttttaa taaccactca taaatcctca tagagtattt gttttcaaaa gacttaacat 3241 gttccagatt atattttatg aattttttta actggaaaag ataaggcaat atctcttcac 3301 taaaaactaa ttctaatttt tcgcttgaga acttggcata gtttgtccac tggaaaatct 3361 caaagccttt aaccaaagga ttcctgattt ccacagttct cgtcatcagc tctctggttg 3421 ctttagctaa tacaccataa gcattttccc tactgatgtt catcatctga gcgtattggt 3481 tataagtgaa cgataccgtc cgttctttcc ttgtagggtt ttcaatcgtg gggttgagta 3541 gtgccacaca gcataaaatt agcttggttt catgctccgt taagtcatag cgactaatcg 3601 ctagttcatt tgctttgaaa acaactaatt cagacataca tctcaattgg tctaggtgat 3661 tttaatcact ataccaattg agatgggcta gtcaatgata attactagtc cttttcccgg 3721 gtgatctggg tatctgtaaa ttctgctaga cctttgctgg aaaacttgta aattctgcta 3781 gaccctctgt aaattccgct agacctttgt gtgttttttt tgtttatatt caagtggtta 3841 taatttatag aataaagaaa gaataaaaaa agataaaaag aatagatccc agccctgtgt 3901 ataactcact actttagtca gttccgcagt attacaaaag gatgtcgcaa acgctgtttg 3961 ctcctctaca aaacagacct taaaacccta aaggcttaag tagcaccctc gcaagctcgg 4021 gcaaatcgct gaatattcct tttgtctccg accatcaggc acctgagtcg ctgtcttttt 4081 cgtgacattc agttcgctgc gctcacggct ctggcagtga atgggggtaa atggcactac 4141 aggcgccttt tatggattca tgcaaggaaa ctacccataa tacaagaaaa gcccgtcacg 4201 ggcttctcag ggcgttttat ggcgggtctg ctatgtggtg ctatctgact ttttgctgtt 4261 cagcagttcc tgccctctga ttttccagtc tgaccacttc ggattatccc gtgacaggtc 4321 attcagactg gctaatgcac ccagtaaggc agcggtatca tcaacaggct tacccgtctt 4381 actgtcccta gtgcttggat tctcaccaat aaaaaacgcc cggcggcaac cgagcgttct 4441 gaacaaatcc agatggagtt ctgaggtcat tactggatct atcaacagga gtccaagcga 4501 gctcgatatc aaattacgcc ccgccctgcc actcatcgca gtactgttgt aattcattaa 4561 gcattctgcc gacatggaag ccatcacaaa cggcatgatg aacctgaatc gccagcggca 4621 tcagcacctt gtcgccttgc gtataatatt tgcccatggt gaaaacgggg gcgaagaagt 4681 tgtccatatt ggccacgttt aaatcaaaac tggtgaaact cacccaggga ttggctgaga 4741 cgaaaaacat attctcaata aaccctttag ggaaataggc caggttttca ccgtaacacg 4801 ccacatcttg cgaatatatg tgtagaaact gccggaaatc gtcgtggtat tcactccaga 4861 gcgatgaaaa cgtttcagtt tgctcatgga aaacggtgta acaagggtga acactatccc 4921 atatcaccag ctcaccgtct ttcattgcca tacgaaattc cggatgagca ttcatcaggc 4981 gggcaagaat gtgaataaag gccggataaa acttgtgctt atttttcttt acggtcttta 5041 aaaaggccgt aatatccagc tgaacggtct ggttataggt acattgagca actgactgaa 5101 atgcctcaaa atgttcttta cgatgccatt gggatatatc aacggtggta tatccagtga 5161 tttttttctc cattttagct tccttagctc ctgaaaatct cgataactca aaaaatacgc 5221 ccggtagtga tcttatttca ttatggtgaa agttggaacc tcttacgtgc cgatcaacgt 5281 ctcattttcg ccagatatc // ]]></de:content> <de:fileName>pj5_00001.gb</de:fileName> </de:sequenceFile> <de:sequenceFile id="1383621055624480"> <de:format>Genbank</de:format> <de:content><![CDATA[LOCUS BMC_nterm_sig_pe 102 bp DNA linear 26-FEB-2010 DEFINITION promoter seq from pBAD33. ACCESSION unknown SOURCE ORGANISM FEATURES Location/Qualifiers misc_feature 55..102 /label=gly_ser_linker CDS 1..102 /label=GFPuv misc_feature 4..54 /label=Clostridium_BMC_sig_pep BASE COUNT 33 a 14 c 29 g 26 t ORIGIN 1 atggaaaata acgctttatt agaacaaata atcaatgaag ttttaaaaaa tatgggtggc 61 agtggtagcg ggagctcggg tggctcaggc tctggttcca gt // ]]></de:content> <de:fileName>BMC_nterm_sig_pep_GFPuv.gb</de:fileName> </de:sequenceFile> <de:sequenceFile id="1383621055625485"> <de:format>Genbank</de:format> <de:content><![CDATA[LOCUS ccmN_nterm_sig_p 108 bp DNA linear 26-FEB-2010 DEFINITION promoter seq from pBAD33. ACCESSION unknown SOURCE ORGANISM FEATURES Location/Qualifiers misc_feature 4..60 /label=ccmN_sig_pep misc_feature 61..108 /label=gly_ser_linker CDS 1..108 /label=GFPuv BASE COUNT 20 a 25 c 40 g 23 t ORIGIN 1 atgaaggtct acggcaagga acagtttttg cggatgcgcc agagcatgtt ccccgatcgc 61 ggtggcagtg gtagcgggag ctcgggtggc tcaggctctg gttccagt // ]]></de:content> <de:fileName>ccmN_nterm_sig_pep_GFPuv.gb</de:fileName> </de:sequenceFile> <de:sequenceFile id="1383621055625791"> <de:format>FASTA</de:format> <de:content><![CDATA[>ssrA_tag_enhance GCGGCGAACGATGAAAACTATAACTATGCGCTGGCGGCG ]]></de:content> <de:fileName>ssrA_tag_enhance.fas</de:fileName> </de:sequenceFile> <de:sequenceFile id="1383621055627870"> <de:format>jbei-seq</de:format> <de:content><![CDATA[<?xml version="1.0" encoding="UTF-8"?> <seq:seq xmlns:seq="http://jbei.org/sequence" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://jbei.org/sequence seq.xsd" > <seq:name>ssrA_tag_GFPuv</seq:name> <seq:circular>false</seq:circular> <seq:sequence>gcggcgaacgatgaaaactatgcgctggcggcg</seq:sequence> <seq:features> <seq:feature> <seq:label>ssrA tag</seq:label> <seq:complement>false</seq:complement> <seq:type>misc_feature</seq:type> <seq:location> <seq:genbankStart>1</seq:genbankStart> <seq:end>33</seq:end> </seq:location> <seq:attribute name="vntifkey" quoted="true" >21</seq:attribute> <seq:seqHash>0d5535e13cc9708d0ff0289af2fae27e564b6bcbcd9242f5140d96957744a517</seq:seqHash> </seq:feature> <seq:feature> <seq:label>GFPuv</seq:label> <seq:complement>false</seq:complement> <seq:type>CDS</seq:type> <seq:location> <seq:genbankStart>1</seq:genbankStart> <seq:end>33</seq:end> </seq:location> <seq:attribute name="vntifkey" quoted="true" >4</seq:attribute> <seq:seqHash>0d5535e13cc9708d0ff0289af2fae27e564b6bcbcd9242f5140d96957744a517</seq:seqHash> </seq:feature> </seq:features> </seq:seq> ]]></de:content> <de:fileName>ssrA_tag_GFPuv.xml</de:fileName> </de:sequenceFile> </de:sequenceFiles> <de:partVOs> <de:partVO id="1318886361742366"> <de:name>pS8c-vector_backbone</de:name> <de:revComp>false</de:revComp> <de:startBP>2016</de:startBP> <de:stopBP>1238</de:stopBP> <de:sequenceFileID>1383621055622750</de:sequenceFileID> <de:parts> <de:part id="1318886361742634"> <de:fas/> </de:part> </de:parts> </de:partVO> <de:partVO id="1318886361747742"> <de:name>BMC_nterm_sig_pep</de:name> <de:revComp>false</de:revComp> <de:startBP>4</de:startBP> <de:stopBP>54</de:stopBP> <de:sequenceFileID>1383621055624480</de:sequenceFileID> <de:parts> <de:part id="1318886361747000"> <de:fas/> </de:part> </de:parts> </de:partVO> <de:partVO id="1318886361752995"> <de:name>ccmN_nterm_sig_pep</de:name> <de:revComp>false</de:revComp> <de:startBP>4</de:startBP> <de:stopBP>60</de:stopBP> <de:sequenceFileID>1383621055625485</de:sequenceFileID> <de:parts> <de:part id="1318886361752939"> <de:fas/> </de:part> </de:parts> </de:partVO> <de:partVO id="1318886361759464"> <de:name>long_gly_ser_linker</de:name> <de:revComp>false</de:revComp> <de:startBP>55</de:startBP> <de:stopBP>102</de:stopBP> <de:sequenceFileID>1383621055624480</de:sequenceFileID> <de:parts> <de:part id="1318886361759096"> <de:fas/> </de:part> </de:parts> </de:partVO> <de:partVO id="1318886361765194"> <de:name>short_gly_ser_linker</de:name> <de:revComp>false</de:revComp> <de:startBP>55</de:startBP> <de:stopBP>78</de:stopBP> <de:sequenceFileID>1383621055624480</de:sequenceFileID> <de:parts> <de:part id="1318886361765102"> <de:fas/> </de:part> </de:parts> </de:partVO> <de:partVO id="1318886361772274"> <de:name>GFPuv</de:name> <de:revComp>false</de:revComp> <de:startBP>1242</de:startBP> <de:stopBP>1952</de:stopBP> <de:sequenceFileID>1383621055622750</de:sequenceFileID> <de:parts> <de:part id="1318886361772744"> <de:fas/> </de:part> </de:parts> </de:partVO> <de:partVO id="1320793316351870"> <de:name>ssrA_tag_5prime</de:name> <de:revComp>false</de:revComp> <de:startBP>1</de:startBP> <de:stopBP>15</de:stopBP> <de:sequenceFileID>1383621055627870</de:sequenceFileID> <de:parts> <de:part id="1320793316351783"> <de:fas/> </de:part> </de:parts> </de:partVO> <de:partVO id="1320793317318921"> <de:name>ssrA_tag_enhanced_5prime</de:name> <de:revComp>false</de:revComp> <de:startBP>1</de:startBP> <de:stopBP>21</de:stopBP> <de:sequenceFileID>1383621055625791</de:sequenceFileID> <de:parts> <de:part id="1320793317318572"> <de:fas>Embed_in_primer_reverse</de:fas> </de:part> </de:parts> </de:partVO> <de:partVO id="1320793489598076"> <de:name>ssrA_tag_3prime</de:name> <de:revComp>false</de:revComp> <de:startBP>16</de:startBP> <de:stopBP>33</de:stopBP> <de:sequenceFileID>1383621055627870</de:sequenceFileID> <de:parts> <de:part id="1320793489598961"> <de:fas>Embed_in_primer_forward</de:fas> </de:part> </de:parts> </de:partVO> </de:partVOs> <de:eugeneRules> <de:eugeneRule> <de:name>Rule_1</de:name> <de:negationOperator>false</de:negationOperator> <de:operand1ID>1318886361765194</de:operand1ID> <de:compositionalOperator>THEN</de:compositionalOperator> <de:operand2ID>1318886361747742</de:operand2ID> </de:eugeneRule> </de:eugeneRules> <de:j5Collection> <de:isCircular>true</de:isCircular> <de:j5Bins> <de:j5Bin id="1383621045334575"> <de:binName>vector_backbone</de:binName> <de:iconID>origin-of-replication</de:iconID> <de:direction>forward</de:direction> <de:dsf>false</de:dsf> <de:fro>2</de:fro> <de:binItems> <de:partID>1318886361742634</de:partID> </de:binItems> </de:j5Bin> <de:j5Bin id="1383621055637671"> <de:binName>nterm_sig_pep</de:binName> <de:iconID>cds</de:iconID> <de:direction>forward</de:direction> <de:dsf>true</de:dsf> <de:binItems> <de:partID>1318886361747000</de:partID> <de:partID>1318886361752939</de:partID> </de:binItems> </de:j5Bin> <de:j5Bin id="1383621055643565"> <de:binName>gly_ser_linker</de:binName> <de:iconID>cds</de:iconID> <de:direction>forward</de:direction> <de:dsf>false</de:dsf> <de:binItems> <de:partID>1318886361759096</de:partID> <de:partID>1318886361765102</de:partID> </de:binItems> </de:j5Bin> <de:j5Bin id="1383621055648427"> <de:binName>GFPuv</de:binName> <de:iconID>cds</de:iconID> <de:direction>forward</de:direction> <de:dsf>false</de:dsf> <de:binItems> <de:partID>1318886361772744</de:partID> </de:binItems> </de:j5Bin> <de:j5Bin id="1383621055654640"> <de:binName>ssrA_tag_5prime</de:binName> <de:iconID>protein-stability-element</de:iconID> <de:direction>forward</de:direction> <de:dsf>true</de:dsf> <de:binItems> <de:partID>1320793316351783</de:partID> <de:partID>1320793317318572</de:partID> </de:binItems> </de:j5Bin> <de:j5Bin id="1383621055659603"> <de:binName>ssrA_tag_3prime</de:binName> <de:iconID>protein-stability-element</de:iconID> <de:direction>forward</de:direction> <de:dsf>false</de:dsf> <de:binItems> <de:partID>1320793489598961</de:partID> </de:binItems> </de:j5Bin> </de:j5Bins> </de:j5Collection> </de:design> Here is the stylized XML schema for the DeviceEditor design file format: <?xml version="1.0" encoding="UTF-8"?> <xs:schema xmlns:xs="http://www.w3.org/2001/XMLSchema" targetNamespace="http://jbei.org/device_editor" xmlns:de="http://jbei.org/device_editor" elementFormDefault="qualified"> <xs:element name="design"> <xs:complexType> <xs:sequence> <xs:element ref="de:version" /> <xs:element ref="de:sequenceFiles" /> <xs:element ref="de:partVOs" /> <xs:element ref="de:eugeneRules" /> <xs:element ref="de:j5Collection"/> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="version"> <xs:simpleType> <xs:restriction base="xs:string"> <xs:pattern value="4.4" /> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="sequenceFiles"> <xs:complexType> <xs:sequence> <xs:element ref="de:sequenceFile" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="sequenceFile"> <xs:complexType> <xs:sequence> <xs:element ref="de:format" /> <xs:element ref="de:content" /> <xs:element ref="de:fileName" /> <xs:element ref="de:icePartId" minOccurs="0" /> <xs:element ref="de:iceEntryURI" minOccurs="0" /> </xs:sequence> <xs:attribute name="id" type="xs:string" /> </xs:complexType> </xs:element> <xs:element name="format"> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="Genbank" /> <xs:enumeration value="FASTA" /> <xs:enumeration value="jbei-seq" /> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="content" type="xs:string" /> <xs:element name="fileName" type="xs:string" /> <xs:element name="icePartId" type="xs:string" /> <xs:element name="iceEntryURI" type="xs:string" /> <xs:element name="partVOs"> <xs:complexType> <xs:sequence> <xs:element ref="de:partVO" minOccurs="0" maxOccurs="unbounded" /> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="partVO"> <xs:complexType> <xs:sequence> <xs:element ref="de:name" /> <xs:group ref="de:seqInfo" minOccurs="0" /> <xs:element ref="de:parts" /> </xs:sequence> <xs:attribute name="id" type="xs:string" /> </xs:complexType> </xs:element> <xs:element name="name" type="xs:string" /> <xs:group name="seqInfo"> <xs:sequence> <xs:element ref="de:revComp" /> <xs:element ref="de:startBP" /> <xs:element ref="de:stopBP" /> <xs:element ref="de:fivePrimeInternalPreferredOverhangs" minOccurs="0"/> <xs:element ref="de:threePrimeInternalPreferredOverhangs" minOccurs="0"/> <xs:element ref="de:sequenceFileID" /> </xs:sequence> </xs:group> <xs:element name="revComp" type="xs:boolean" /> <xs:element name="startBP" type="xs:positiveInteger" /> <xs:element name="stopBP" type="xs:positiveInteger" /> <xs:element name="fivePrimeInternalPreferredOverhangs" type="xs:string" /> <xs:element name="threePrimeInternalPreferredOverhangs" type="xs:string" /> <xs:element name="sequenceFileID" type="xs:string" /> <xs:element name="parts"> <xs:complexType> <xs:sequence> <xs:element ref="de:part" maxOccurs="unbounded" /> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="part"> <xs:complexType> <xs:sequence> <xs:element ref="de:fas" /> </xs:sequence> <xs:attribute name="id" type="xs:string" /> </xs:complexType> </xs:element> <xs:element name="fas"> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="" /> <xs:enumeration value="DIGEST" /> <xs:enumeration value="Direct Synthesis" /> <xs:enumeration value="PCR" /> <xs:enumeration value="Embed_in_primer_reverse" /> <xs:enumeration value="Embed_in_primer_forward" /> <xs:enumeration value="Annealed Oligos"></xs:enumeration> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="eugeneRules"> <xs:complexType> <xs:sequence> <xs:element ref="de:eugeneRule" minOccurs="0" maxOccurs="unbounded" /> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="eugeneRule"> <xs:complexType> <xs:sequence> <xs:element ref="de:name" /> <xs:element ref="de:negationOperator" /> <xs:element ref="de:operand1ID" /> <xs:element ref="de:compositionalOperator" /> <xs:group ref="de:operand2" /> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="negationOperator" type="xs:boolean" /> <xs:element name="operand1ID" type="xs:string" /> <xs:element name="compositionalOperator"> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="AFTER" /> <xs:enumeration value="BEFORE" /> <xs:enumeration value="WITH" /> <xs:enumeration value="THEN" /> <xs:enumeration value="NEXTTO" /> <xs:enumeration value="MORETHAN" /> </xs:restriction> </xs:simpleType> </xs:element> <xs:group name="operand2"> <xs:choice> <xs:element ref="de:operand2ID" /> <xs:element ref="de:operand2Number" /> </xs:choice> </xs:group> <xs:element name="operand2ID" type="xs:string" /> <xs:element name="operand2Number" type="xs:nonNegativeInteger" /> <xs:element name="j5Collection"> <xs:complexType> <xs:sequence> <xs:element ref="de:isCircular" /> <xs:element ref="de:j5Bins" /> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="isCircular" type="xs:boolean" /> <xs:element name="j5Bins"> <xs:complexType> <xs:sequence> <xs:element ref="de:j5Bin" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="j5Bin"> <xs:complexType> <xs:sequence> <xs:element ref="de:binName" /> <xs:element ref="de:iconID" /> <xs:element ref="de:direction" /> <xs:element ref="de:dsf" /> <xs:element ref="de:fro" minOccurs="0" /> <xs:element ref="de:extra5PrimeBps" minOccurs="0" /> <xs:element ref="de:extra3PrimeBps" minOccurs="0" /> <xs:element ref="de:binItems" /> </xs:sequence> <xs:attribute name="id" type="xs:string" /> </xs:complexType> </xs:element> <xs:element name="binName" type="xs:string" /> <xs:element name="iconID"> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="assembly-scar" /> <xs:enumeration value="blunt-restriction-site" /> <xs:enumeration value="cds" /> <xs:enumeration value="five-prime-overhang" /> <xs:enumeration value="five-prime-sticky-restriction-site" /> <xs:enumeration value="insulator" /> <xs:enumeration value="operator" /> <xs:enumeration value="origin-of-replication" /> <xs:enumeration value="primer-binding-site" /> <xs:enumeration value="promoter" /> <xs:enumeration value="protease-site" /> <xs:enumeration value="protein-stability-element" /> <xs:enumeration value="restriction-enzyme-recognition-site" /> <xs:enumeration value="ribonuclease-site" /> <xs:enumeration value="ribosome-entry-site" /> <xs:enumeration value="rna-stability-element" /> <xs:enumeration value="signature" /> <xs:enumeration value="terminator" /> <xs:enumeration value="three-prime-overhang" /> <xs:enumeration value="three-prime-sticky-restriction-site" /> <xs:enumeration value="user-defined" /> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="direction"> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="forward" /> <xs:enumeration value="reverse" /> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="dsf" type="xs:boolean" /> <xs:element name="fro" type="string" /> <xs:element name="extra5PrimeBps" type="xs:int" /> <xs:element name="extra3PrimeBps" type="xs:int" /> <xs:element name="binItems"> <xs:complexType> <xs:sequence> <xs:element ref="de:partID" minOccurs="0" maxOccurs="unbounded" /> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="partID" type="xs:string" /> </xs:schema> |
||||||