| 5 |
The BioBrick approach |
| BioBricks standardize the DNA assembly process, facilitating automation and part re-use. There are several BioBrick assembly standards, such as that originally developed at MIT (Shetty 2008), as well as the UC Berkeley BglBrick standard (Anderson 2010), which is depicted in the example shown below. BglBrick assembly of partA, partB, and the partA-bearing vector backbone: 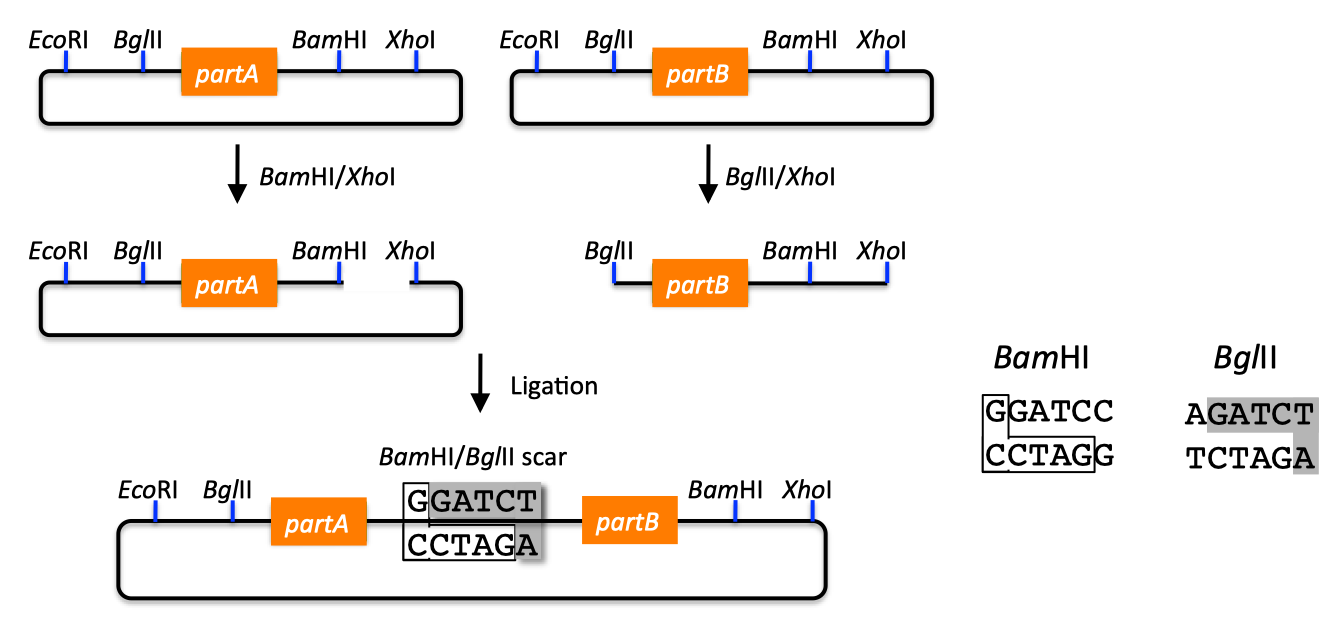 In the BglBrick standard, a part (or DNA sequence fragment that is nominally associated with a biological function) is flanked with two restriction enzyme sites at its 5' end, namely EcoRI and BglII, and is flanked with BamHI and XhoI at its 3' terminus. To comply with the BglBrick standard, these four restriction sites must be absent from the sequence of the part itself. The "BglBrick", then, spans from the EcoRI to the XhoI site, and the BioBrick-bearing vector backbone makes up the residual plasmid sequence, which should also be devoid of the four BglBrick restriction sites. To assemble partA followed by partB, followed by the partA-bearing vector backbone, the partA BglBrick vector is digested with BamHI and XhoI, and the partB vector is digested with BglII and XhoI. The resulting digest fragments containing partA and partB are then ligated together, resulting in the desired plasmid. The overhang sequences resulting from BamHI and BglII digest are complementary (base-pair/anneal perfectly with one-another), but the resulting ligation product sequence is not recognized/recleaved by either BamHI or BglII. Thus, the assembly results in a new BglBrick, containing partA followed by a six bp scar sequence, followed by partB. A key consequence of BglBrick assembly is that assembling two parts results in a new BglBrick, so that this process can be iterated successively to assemble an arbitrary number of parts together, using the same protocol repetitively. It is possible to assemble partB in front of partA, and/or to select the partA or partB-bearing vector backbone for the resulting construct, by using different combinations of the four BglBrick restriction enzymes. Other BioBrick standards are completely analogous to BglBricks, and simply use alternate sets of the four restriction enzymes. Contrasting with the traditional approach, there are several advantages to using BioBricks: 1) only four restriction enzymes are utilized, 2) once a part is BioBrick'd (i.e. domesticated), it is never necessary to re-clone it (or even re-PCR amplify it, reducing the probability of PCR-derived mutations), and 3) assembling an arbitrary number of parts (in any desired arrangement) is no more difficult than putting two together (plasmid size considerations aside). It should be highlighted that, in contrast with SLIC, Gibson, CPEC, SLiCE and Golden Gate methods, BioBrick assembly not only standardizes the assembly process (e.g. the set of four restriction enzymes, protocols, etc.), but also physically standardizes the BioBrick'd parts themselves, as they all have the same 5' and 3' terminal sequences, and internally share the same 6-bp scar vestiges of prior assemblies. There are burgeoning repositories of these standardized parts (physical and/or informatic), such as the iGEM Registry of Standard Biological Parts, and supporting organizations, such as the BioBricks Foundation, that allow and facilitate researcher re-use of characterized and validated parts, preempting wasteful redundant efforts. Returning to the previous DNA assembly challenge, here's how we could use BioBrick assembly to put together the pathway: 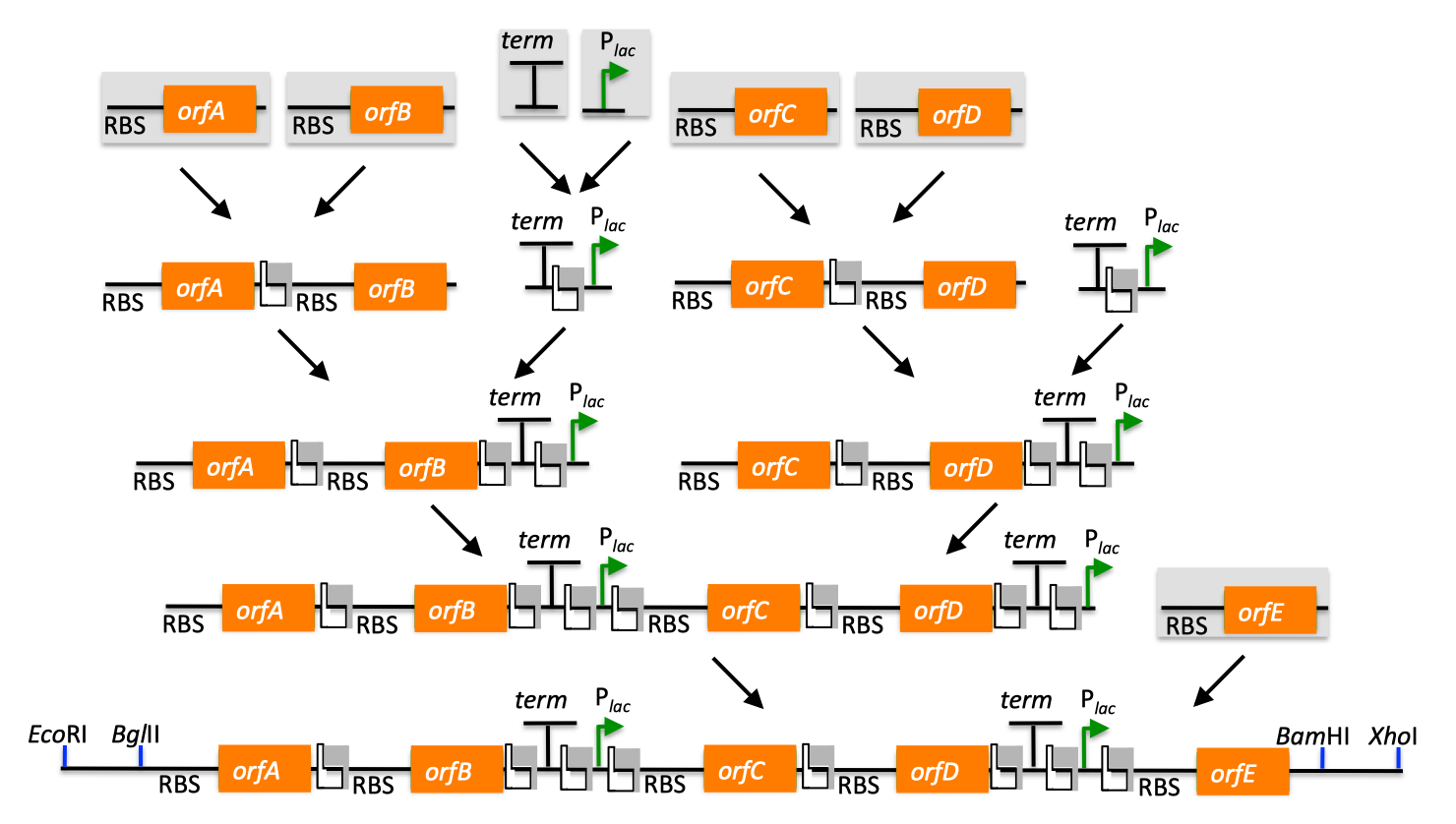 Note that there are many different possible routes (assembly trees) to put together this pathway using BioBricks. Some of the intermediate parts, such as the terminator fused to the promoter, need only be made once, and can be re-used multiple times. Recently, algorithms have been developed (Densmore 2010) to optimize the design of binary BioBrick assembly trees, and an automated in vivo BglBrick assembly process utilizing liquid-handling robotics has been demonstrated (Leguia 2011). BioBrick limitations and obstacles Despite the many merits of the BioBrick approach, there are some drawbacks. First, there is no control over the existence and sequence of the 6-bp scars resulting from each binary BioBrick assembly. While in many cases, these scars will not prove problematic, there are scenarios where the scar sequences, affecting coding sequences or mRNA secondary structure, can adversely perturb the desired protein, RBS, terminator, etc., function. Second, unless every intermediate part is archived along the binary assembly tree, it is necessary to repeat the entire process from scratch in order to replace a composite part (e.g. orfC in the example above) in the assembled BioBrick; even if all intermediate parts are archived, many of the steps must still be repeated. Third, combinatorial library diversity generation is potentially at odds with the BioBrick assembly process, because diversity must be recaptured after each and every binary assembly step (which requires aggregating 5 times as many post-transformation colonies per binary assembly as the sequence diversity to be retained). Fourth, BioBrick assembly only works with previously BioBrick'd parts, and another cloning method must be used to first create the BioBricks to be assembled. Given the availability of design tools for BioBrick assembly, and an automated in-vivo BioBrick assembly methodology, j5 does not design BioBrick assembly, but instead designs for the SLIC, Gibson, CPEC, SLiCE and Golden Gate methods, which address the short comings of the BioBrick approach, and retain compatibility with the established BioBrick standard(s) (i.e. the methods employed by j5 can be used to assemble BioBrick vectors, although not using BioBrick assembly to do so). |
|||||