| 32 |
Sequences list file and zipped sequences file |
| The sequences list file is a CSV file that contains a list of all of the source/template sequences from which DNA parts will be defined. The first line in the file is a header line that does not contain sequence file information, but rather just serves to name the columns below, namely Sequence File Name and Format. Sequence File Name column fields: The file names, including extention (e.g. ".gb", ".fas", or ".xml"). Format column fields: Should either contain "Genbank", "FASTA", "jbei-seq", or "SBOLXML". Here is what an example sequences list file looks like (stylized for clarity): 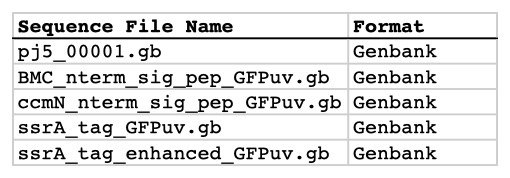 Here is the actual example sequences list CSV file (myseqlist.csv): The zipped sequences file is just a compressed (.zip) file that contains all of the sequence files (in Genbank, FASTA, jbei-seq , or SBOL XML format) listed in the sequences list file. The sequence files may be zipped directly, or as part of a directory structure that contains all of the sequence files. In the latter case, please ensure that no two sequence files within the directory structure have identical names. If there is a repeated sequence file name (at least for those sequence files listed in the sequence list input file), j5 will issue a warning, and will use the first sequence file with a matching name it finds. Here is an example zipped sequences file that corresponds to the example sequences list file above (myseqs.zip): |
||||||